Guanine-Cytosine Content Analysis and Basics of DNA Sequence Statistics
Procedure
Follow ( https://vlab.amrita.edu/index.php?sub=3&brch=311&sim=1835&cnt=2) to install R in personal computer.
Install sequinR package. Follow the code
install.packages('seqinr') library('seqinr')
Procedure to calculate GC content in a personal computer
Follow the code in the command window:
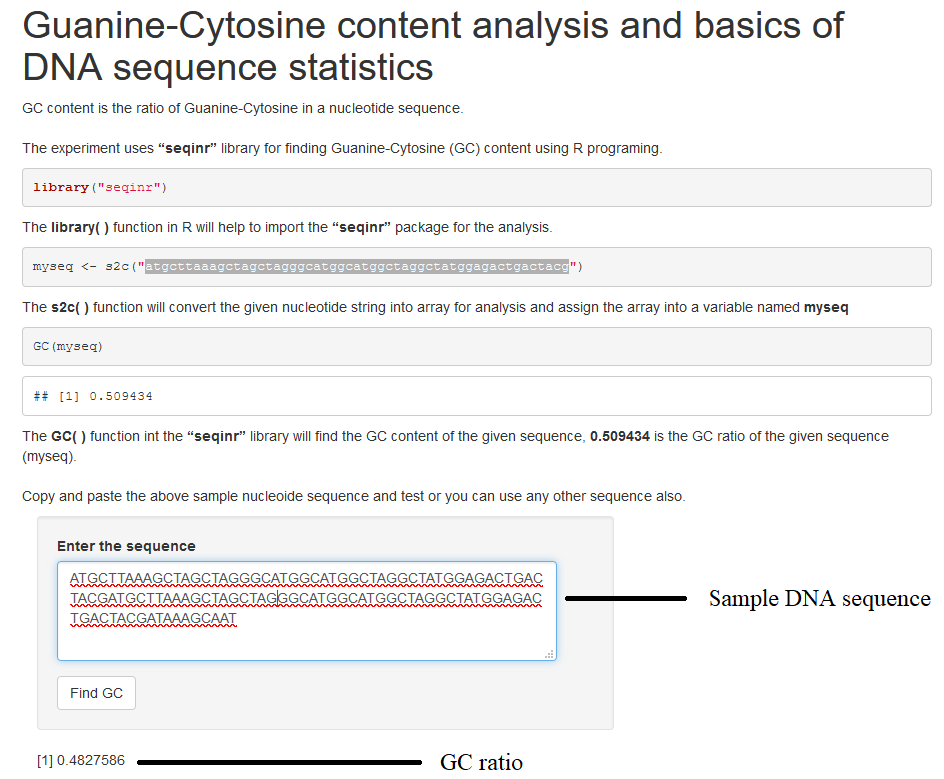
library("seqinr") seq1<-s2c("atgcttaaagctagctagggcatggcatggctaggctatggagactgactacg") GC(seq1)Click execute button for output.
Description
The experiment uses “seqinr” library for finding Guanine-Cytosine (GC) content in R programing. GC content is the ratio of Guanine-Cytosine in a nucleotide sequence. First line of code is importing the seqinR library into the workspace. The function “s2c” convert the string into an array and assign to a variable named seq1 and the GC( ) function in seqinr library calculates the GC content of the given nucleotide sequence.
Procedure to work simulator
- Paste any DNA sequence in the textbox provided. Click on “ Find GC” button
- GC ratio will be displayed in the GUI.