Computing scoring matrices for amino acids and long pairwise alignment in R
Procedure to run the simulator
- User can enter the sequence to be compared in respective “ Sequence 1 and Sequence 2” boxes.
- Retrieve sequence data in FASTA format from NCBI. Consider examples,
Sequence 1 - Keratin[Homosapiens]
AAB59562.1 keratin [Homo sapiens]
MTTCSRQFTSSSSMKGSCGIGGGIGAGSSRISSVLAGGSCRAPNTYGGGLSVSSSRFSSGGAYGLGGGYG
GGFSSSSSSFGSGFGGGYGGGLGAGLGGGFGGGFAGGDGLLVGSEKVTMQNLNDRLASYLDKVRALEEAN
ADLEVKIRDWYQRQRPAEIKDYSPYFKTIEDLRNKILTATVDNANVLLQIDNARLAADDFRTKYETELNL
RMSVEADINGLRRVLDELTLARADLEMQIESLKEELAYLKKNHEEEMNALRGQVGGDVNVEMDAAPGVDL
SRILNEMRDQYEKMAEKNRKDAEEWFFTKTEELNREVATNSELVQSGKSEISELRRTMQNLEIELQSQLS
MKASLENSLEETKGRYCMQLAQIQEMIGSVEEQLAQLRCEMEQQNQEYKILLDVKTRLEQEIATYRRLLE
GEDAHLSSSQFSSGSQSSRDVTSSSRQIRTKVMDVHDGKVVSTHEQVLRTKN
Sequence 2 - keratin [Rattus norvegicus]
AAA41473.1 keratin [Rattus norvegicus]
MDFSRRSFHRSLSSSSQGPALSTSGSLYRKGTMQRLGLHSVYGGWRHGTRISVSKTTMSYGNHLSNGGDL
FGGNEKLAMQNLNDRLASYLEKVRSLEQSNSKLEAQIKQWYETNAPSTIRDYSSYYAQIKELQDQIKDAQ
IENARCVLQIDNAKLAAEDFRLKFETERGMRITVEADLQGLSKVYDDLTLQKTDLEIQIEELNKDLALLK
KEHQEEVEVLRRQLGNNVNVEVDAAPGLNLGEIMNEMRQKYEILAQKNLQEAKEQFERQTQTLEKQVTVN
IEELRGTEVQVTELRRSYQTLEIELQSQLSMKESLERTLEETKARYASQLAAIQEMLSSLEAQLMQIRSD
TERQNQEYNILLDIKTRLEQEIATYRRLLEGEDIKTTEYQLNTLEAKDIKKTRKIKTVVEEVVDGKVVSS
EVKEIEENI
Then click on Align Seq tab to run simulator.
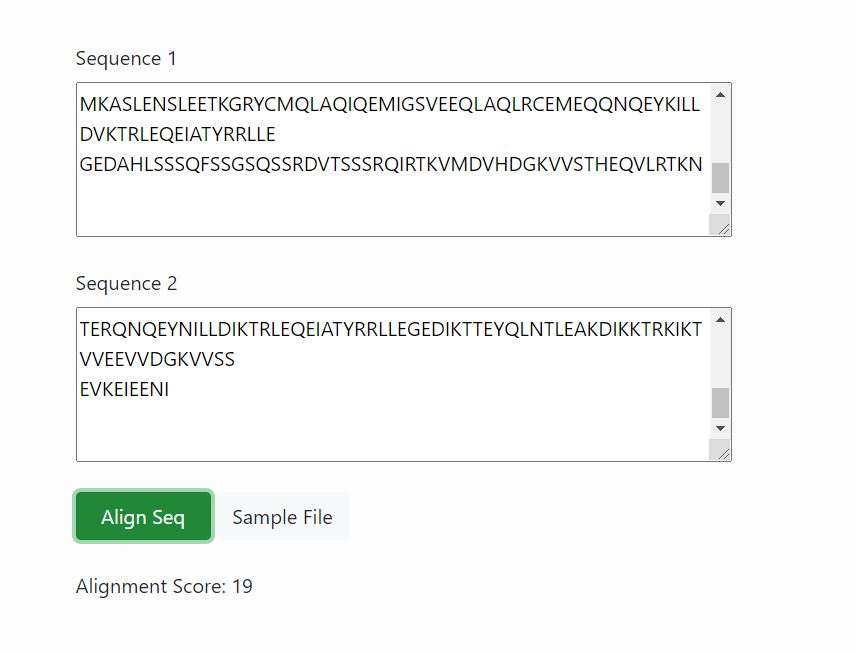
The alignment score for the given aminoacid sequence matrix is given as output which indicates the relative score obtained by matching two characters in a sequence alignment.
User can also download simple file given in the GUI and follow Steps 1 and 2 to get output.
DIY
Follow ( https://vlab.amrita.edu/index.php?sub=3&brch=311&sim=1835&cnt=2) to install R in personal computer.
Install the SeqinR package.
To load “SeqinR” R package follow
library("seqinr")
Import “seqinr” library to R workspace
Connect to the ACNUC database
Query the sequences using accession number and assign to a variable
Import “Biostrings” library to R workspace
Load Blosum50 scoring matrix
Convert the amino acid sequences into strings
Convert the string into uppercase
Pairwise align the sequences with gap values input
Assign the alignment data into a matrix
